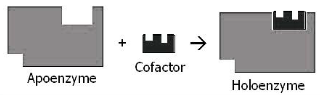
To aid in catalytic activity, many enzymes require the inclusion of a tiny molecule known as a cofactor. A cofactor is a non-protein molecule that performs chemical reactions that the usual 20 amino acids cannot. Cofactors can be inorganic (metals) or organic (small organic compounds) (coenzymes). Cofactors, which are typically coenzymes or metal ions , are inorganic and organic molecules that help enzymes perform their functions. Coenzymes are nonprotein organic molecules that bind to apoenzyme protein molecules to produce active holoenzymes. They are mostly vitamin derivatives that are soluble in water due to phosphorylation. Holoenzyme refers to apoenzyme in the presence of its cofactor. A holoenzyme is an enzyme that is both full and catalytically active. The majority of cofactors are firmly bonded rather than covalently bound. Organic prosthetic groups, such as iron ions or vitamins, can, however, be covalently bound. DNA polymerase and RNA polymerase are examples of holoenzymes, which comprise several protein components. All of the subunits required for activity are found in the full complexes.
Holoenzyme
The term holoenzyme refers to an apoenzyme that is complete and catalytically active, as well as the cofactor. A cofactor might be a tiny organic molecule or a metal ion. The majority of metal ions are covalently or noncovalently linked to the enzyme. Prosthetic groupings are what they’re called. Coenzymes are a type of small chemical molecule.
Coenzymes can bind to enzymes either tightly or loosely. Cu, Co, Mg, Mn, Ni, Fe and other ions can be used as prosthetic groups. NADP, NAD, FAD, folic acid, biotin, and other coenzymes are examples. Cofactors bind to the enzyme succinate dehydrogenase. DNA polymerase and RNA polymerase are two examples of holoenzymes. Multi-protein subunits make up these enzymes. As a result, they are both complete and complex. A biological reaction can only be catalysed by holoenzymes.
Examples of Holoenzyme
DNA Polymerase: The polymerisation of deoxyribonucleotides into a DNA strand is catalysed by DNA polymerase, a holoenzyme. DNA polymerase is a vital component of DNA replication. It employs the undamaged DNA strand as a template for constructing the new strand. The newly polymerised DNA strand is similar to the template’s original partner strand and complementary to the template strand. The catalytic activity of DNA polymerase is catalysed by the magnesium ion.
RNA Polymerase: is also an RNA-catalysing holoenzyme. Transcription, the process of creating RNA chains using DNA genes as templates, requires RNA polymerase. At the 3′ end of an RNA transcript, it polymerizes ribonucleotides.
Difference Between Apoenzyme and Holoenzyme
Definition
Apoenzyme: Apoenzyme is the inactive version of an enzyme that activates when a cofactor is bound.
Holoenzyme: Holoenzyme refers to an apoenzyme that is complete and catalytically active, as well as the cofactor.
Activity of Catalysis
Apoenzyme: Apoenzyme is the enzyme’s inactive form.
Holoenzyme: Holoenzyme is the enzyme’s catalytically active form.
Content
Apoenzyme: Apoenzyme consists of the protein part of the enzyme.
Holoenzyme: Holoenzyme consists of the apoenzyme and one or several cofactors.
Complexity
Apoenzyme: are imperfect enzymes with a lower level of complexity.
Holoenzymes: are enzymes that are both complete and complicated.
Examples
Apoenzyme: Apoenzymes are the catalytic components of the DNA polymerase enzyme.
Holoenzyme: The multi-subunit complex of DNA polymerase is referred to as a holoenzyme.
Function of Holoenzyme
The holoenzyme is an enzyme that is ready to perform its catalytic role, which is to speed up chemical reactions that occur in various locations. The functions may differ based on the holoenzyme’s unique action. The DNA polymerase, which ensures that DNA is duplicated correctly, is one of the most crucial.
DNA Polymerase III Holoenzyme
In bacterial DNA replication, is the major enzyme complex involved. In 1970, Thomas Kornberg (son of Arthur Kornberg) and Malcolm Gefter discovered it. The complex has a high processivity (number of nucleotides added per binding event) and, when it comes to the replication of the E.coli genome, collaborates with four other DNA polymerases (Pol I, Pol II, Pol IV, and Pol V).
The DNA Pol III holoenzyme is the vital holoenzyme which is involved in the replication activity, and it also possess proofreading capabilities, that helps them to rectify replication errors using exonuclease activity to read 3’5’ and synthesise 5’3’. The replisome, which is positioned at the replication fork, contains DNA Pol III.
Mechanism of Action of DNA Polymerase III
Base pairs are synthesised at a rate of roughly 1000 nucleotides per second by DNA polymerase III. After strand separation at the replication origin, DNA Pol III activity commences. Because DNA synthesis cannot begin from scratch, primase (an RNA polymerase) creates an RNA primer that is complementary to a portion of the single-stranded DNA:
Addition onto 3’OH
As replication occurs and the replisome advances, DNA polymerase III reaches the RNA primer and starts replicating the DNA, adding to the primer’s 3’OH.
Synthesis of DNA
Depending on whether this occurs on the leading or lagging strand (Okazaki fragment) of the DNA, DNA polymerase III will next synthesis a continuous or discontinuous strand of DNA. Because DNA polymerase III has a high processivity, it can create DNA fast. The clamps that “hold” onto the DNA strands contribute to the high processivity.
Removal of Primer
The RNA primer is removed by DNA polymerase I via the nick translation process after replication of the appropriate area. DNA ligase can ligate the DNA-DNA nick between the new fragment and the prior strand once the RNA primer is removed. For high fidelity, high-processivity DNA replication, DNA polymerase I and III, as well as a variety of additional enzymes, are required.
Conclusion
The holoenzyme mechanism allows for termination at 6 T nucleotides, although optimal performance requires C53/37 and C11. Furthermore, the holoenzyme process has two components: delaying elongation and terminator arrest prevention, both of which require C11, C53, and C37. The two states of enzymes that catalyse biological reactions inside the cell are apoenzyme and holoenzyme. Apoenzyme is the inactive protein component of the enzyme. When one or more cofactors bind to the apoenzyme, it becomes active. The apoenzyme’s active form is known as the holoenzyme. The structure and catalytic activity of each form of the enzyme are the fundamental differences between apoenzyme and holoenzyme.
 Profile
Profile Settings
Settings Refer your friends
Refer your friends Sign out
Sign out



