The portion of an enzyme that binds substrate molecules is known as the active site. This is critical for the catalytic activity of the enzyme.
Enzymes are proteins that reduce the activation energy of chemical reactions, allowing them to move much faster. They accomplish this by interacting with chemical reactants – the substrates – in ways that encourage them to undergo chemical reactions. This interaction takes place in the enzyme’s active site, where the substrates are bound to maximise their chances of reacting.
Active Site Specificity:
Ionic Charge : Positive and negative charges are attracted to each other. Similar charges, such as two positive charges, repel each other actively. Another way an enzyme active site might attract substrates or parts of substrates while rejecting others to obtain the perfect fit is through a mechanism known as repulsion.
Polarity : Non-polar molecules prefer other non-polar molecules, whereas polar molecules prefer other polar ones. Certain amino acids in the active site might therefore attract or repel different sections of the substrate, resulting in a better match.
Hydrophobicity or hydrophilicity : Opposites do not attract in this scenario. Instead, what attracts what draws. Hydrophilic amino acids attract hydrophilic substrates, while hydrophobic amino acids attract other hydrophobic compounds.
Size and shape of the active site : The active sites of enzymes are structured in such a way that they can only ‘fit’ with specific substrates.
Some vitamins and minerals are necessary because they act as cofactors, assisting enzymes in binding to their substrates. Enzymes involved in energy production, for example, utilise various B vitamins as cofactors. That’s why B vitamins are found in many energy “shots” and supplements.
The Active Site’s Features
- The active site is a three-dimensional cleft produced by groups from various amino acid sequences.
- The active site of an enzyme occupies a modest portion of the total volume of the enzyme.
- Clefts or crevices are active locations.
- Multiple weak attractions bind substrates to enzymes.
- Binding specificity is determined by the precise arrangement of atoms in an active site.
The Process :
An enzyme will bind to one or more reactant molecules in order to catalyse a process. The enzyme’s substrates are those compounds.
One substrate can be broken down into a number of products in some reactions. In other cases, two substrates are combined to form a bigger molecule or portions are swapped. In fact, there is most likely an enzyme to speed up any biological reaction you can think of!
The active site (where the catalytic “activity” takes place) is the region of the enzyme that binds the substrate.
Proteins are formed up of units called amino acids, and the amino acids that make up the active site in enzymes give it its properties. These amino acids can have large or small side chains, acidic or basic, hydrophilic or hydrophobic side chains.
The active site’s amino acid composition, as well as their relative placements in 3D space, give it a distinct size, shape, and chemical behaviour. An enzyme’s active site is specifically designed to bind to a specific target—the enzyme’s substrate or substrates—and assist them in undergoing a chemical reaction thanks to these amino acids.
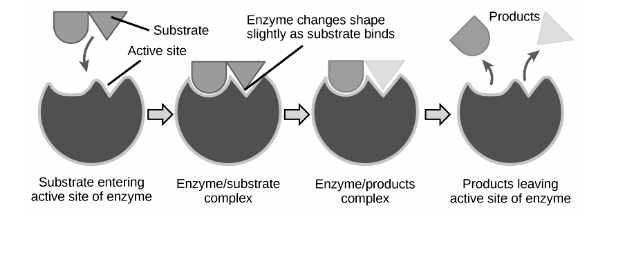
Binding Theories :
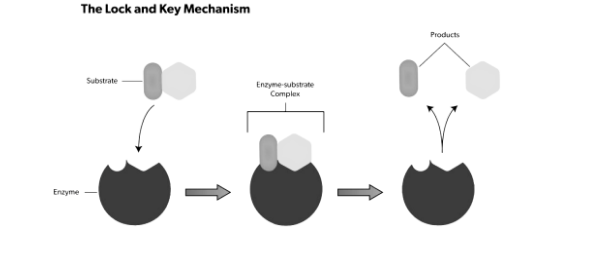
Lock and Key Model :
Emil Fischer, a 19th-century chemist, proposed this concept. He claimed that the active site and substrate are two stable structures that, like a key and a lock, fit together properly without any further change. The interactions between substrates will be strongest if one perfectly binds to its active site, resulting in high catalytic efficiency.
As time passed, the model’s shortcomings became apparent. The competitive enzyme inhibitor methylglucoside, for example, can bind tightly and precisely to the active site of 4-alpha-glucanotransferase. On methylglucoside, however, 4-alpha-glucanotransferase is inactive, and no glycosyl transfer occurs. Because of its strong binding, the Lock and Key hypothesis cannot explain this, as it would predict a high efficiency of methylglucoside glycosyl transfer. Apart from competitive inhibition, this theory is unable to explain the mechanism of non-competitive inhibitors, which do not bind to the active site yet nonetheless influence catalytic activity.
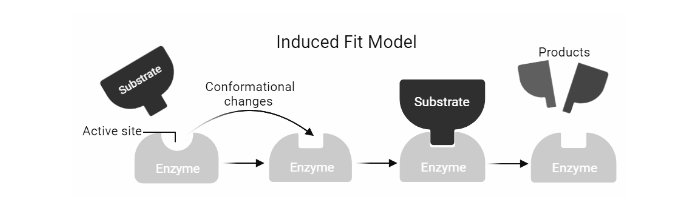
Induced Fit :
The active site and the binding part of the substrate are not exactly complementary, according to Daniel Koshland’s theory of enzyme-substrate binding. The induced fit model is a variation on the lock-and-key model, in which an active site is flexible and changes form until the substrate is completely bound. This model works in the same way that a glove does: the glove changes shape to fit the hand. The enzyme’s shape attracts its substrate at first. The surface of an enzyme is flexible, and only the right catalyst may cause interaction, which leads to catalysis. As the substrate is bound, conformational changes may occur. The reaction products will move away from the enzyme after the reaction, and the active site will revert to its original structure. The fact that the entire protein domain can migrate several nanometers during catalysis supports this notion. This mobility of the protein surface can lead to the formation of microenvironments that are conducive to catalysis.
Conclusion :
The active site is a groove or pocket created by the protein’s folding structure. This region of the enzyme is the only one that binds to the substrate. The enzyme-substrate complex has a three-dimensional structure. Because of this, as well as the chemical features of the amino acids and co-factors, only a single substrate may bind to the site, making it specific to individual proteins.
 Profile
Profile Settings
Settings Refer your friends
Refer your friends Sign out
Sign out




